Model cycling levels as a function of explanatory variables
Source:R/model.R
model_pcycle_pct_2020.RdModel cycling levels as a function of explanatory variables
model_pcycle_pct_2020(pcycle, distance, gradient, weights)Arguments
Examples
# l = get_pct_lines(region = "isle-of-wight")
# l = get_pct_lines(region = "cambridgeshire")
l = wight_lines_pct
pcycle = l$bicycle / l$all
pcycle_dutch = l$dutch_slc / l$all
m1 = model_pcycle_pct_2020(
pcycle,
distance = l$rf_dist_km,
gradient = l$rf_avslope_perc - 0.78,
weights = l$all
)
m2 = model_pcycle_pct_2020(
pcycle_dutch, distance = l$rf_dist_km,
gradient = l$rf_avslope_perc - 0.78,
weights = l$all
)
m3 = model_pcycle_pct_2020(
pcycle_dutch, distance = l$rf_dist_km,
gradient = l$rf_avslope_perc - 0.78,
weights = rep(1, nrow(l))
)
m1
#>
#> Call: stats::glm(formula = pcycle ~ distance + sqrt(distance) + I(distance^2) +
#> gradient + distance * gradient + sqrt(distance) * gradient,
#> family = "quasibinomial", weights = weights)
#>
#> Coefficients:
#> (Intercept) distance sqrt(distance)
#> -6.79130 -1.04186 4.17349
#> I(distance^2) gradient distance:gradient
#> 0.01768 0.63445 0.03433
#> sqrt(distance):gradient
#> -0.48555
#>
#> Degrees of Freedom: 136 Total (i.e. Null); 130 Residual
#> Null Deviance: 657.4
#> Residual Deviance: 351.3 AIC: NA
plot(l$rf_dist_km, pcycle, cex = l$all / 100, ylim = c(0, 0.5))
points(l$rf_dist_km, m1$fitted.values, col = "red")
points(l$rf_dist_km, m2$fitted.values, col = "blue")
points(l$rf_dist_km, pcycle_dutch, col = "green")
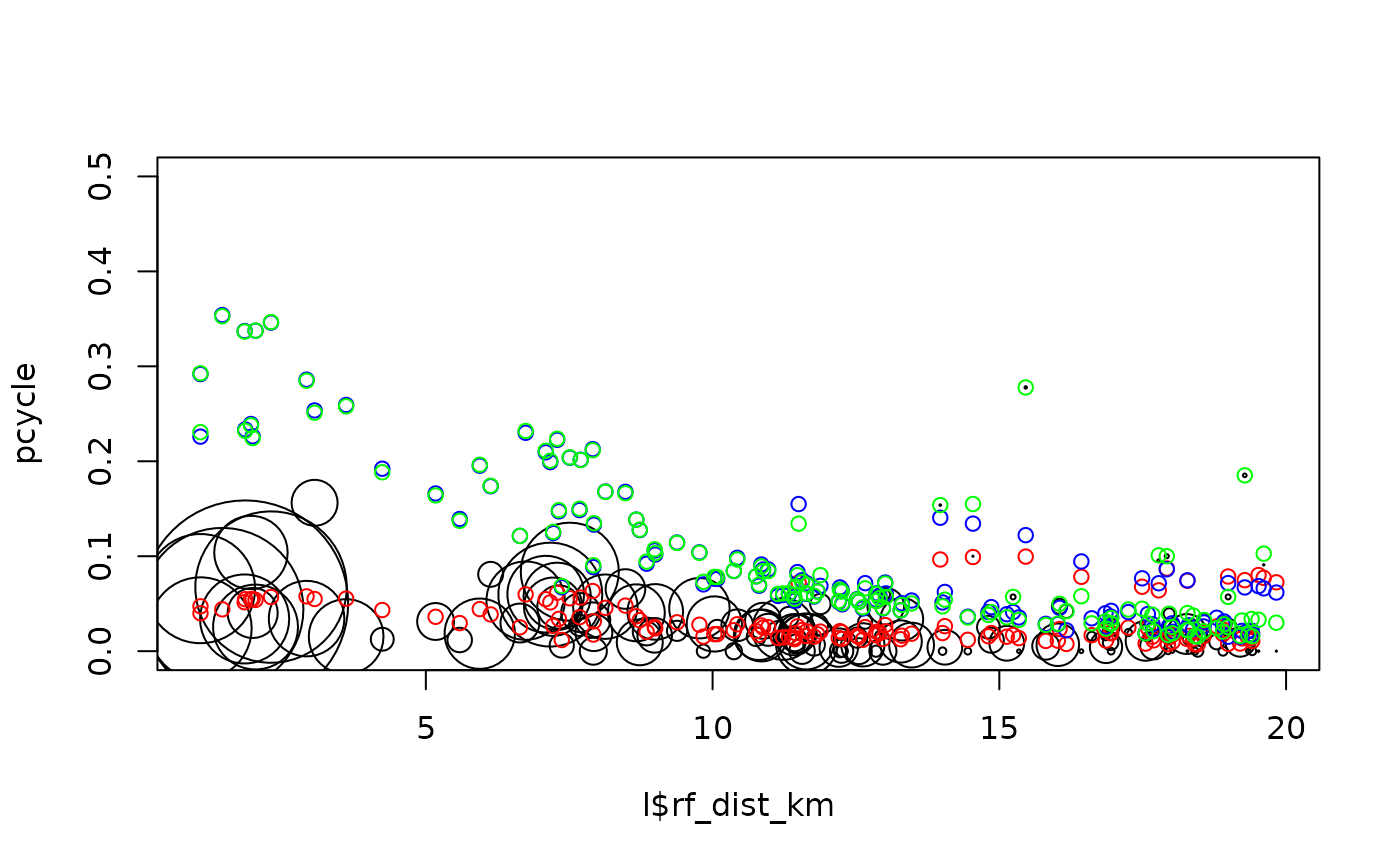 cor(l$dutch_slc, m2$fitted.values * l$all)^2 # 95% captured
#> [1] 0.9998731
# identical means:
mean(l$dutch_slc)
#> [1] 34.18643
mean(m2$fitted.values * l$all)
#> [1] 34.18643
pct_coefficients_2020 = c(
alpha = -4.018 + 2.550,
d1 = -0.6369 -0.08036,
d2 = 1.988,
d3 = 0.008775,
h1 = -0.2555,
i1 = 0.02006,
i2 = -0.1234
)
pct_coefficients_2020
#> alpha d1 d2 d3 h1 i1 i2
#> -1.468000 -0.717260 1.988000 0.008775 -0.255500 0.020060 -0.123400
m2$coef
#> (Intercept) distance sqrt(distance)
#> -1.11820425 -0.63433011 1.61587978
#> I(distance^2) gradient distance:gradient
#> 0.01071045 -0.43192958 -0.03685552
#> sqrt(distance):gradient
#> 0.13203862
plot(pct_coefficients_2020, m2$coeff)
cor(l$dutch_slc, m2$fitted.values * l$all)^2 # 95% captured
#> [1] 0.9998731
# identical means:
mean(l$dutch_slc)
#> [1] 34.18643
mean(m2$fitted.values * l$all)
#> [1] 34.18643
pct_coefficients_2020 = c(
alpha = -4.018 + 2.550,
d1 = -0.6369 -0.08036,
d2 = 1.988,
d3 = 0.008775,
h1 = -0.2555,
i1 = 0.02006,
i2 = -0.1234
)
pct_coefficients_2020
#> alpha d1 d2 d3 h1 i1 i2
#> -1.468000 -0.717260 1.988000 0.008775 -0.255500 0.020060 -0.123400
m2$coef
#> (Intercept) distance sqrt(distance)
#> -1.11820425 -0.63433011 1.61587978
#> I(distance^2) gradient distance:gradient
#> 0.01071045 -0.43192958 -0.03685552
#> sqrt(distance):gradient
#> 0.13203862
plot(pct_coefficients_2020, m2$coeff)
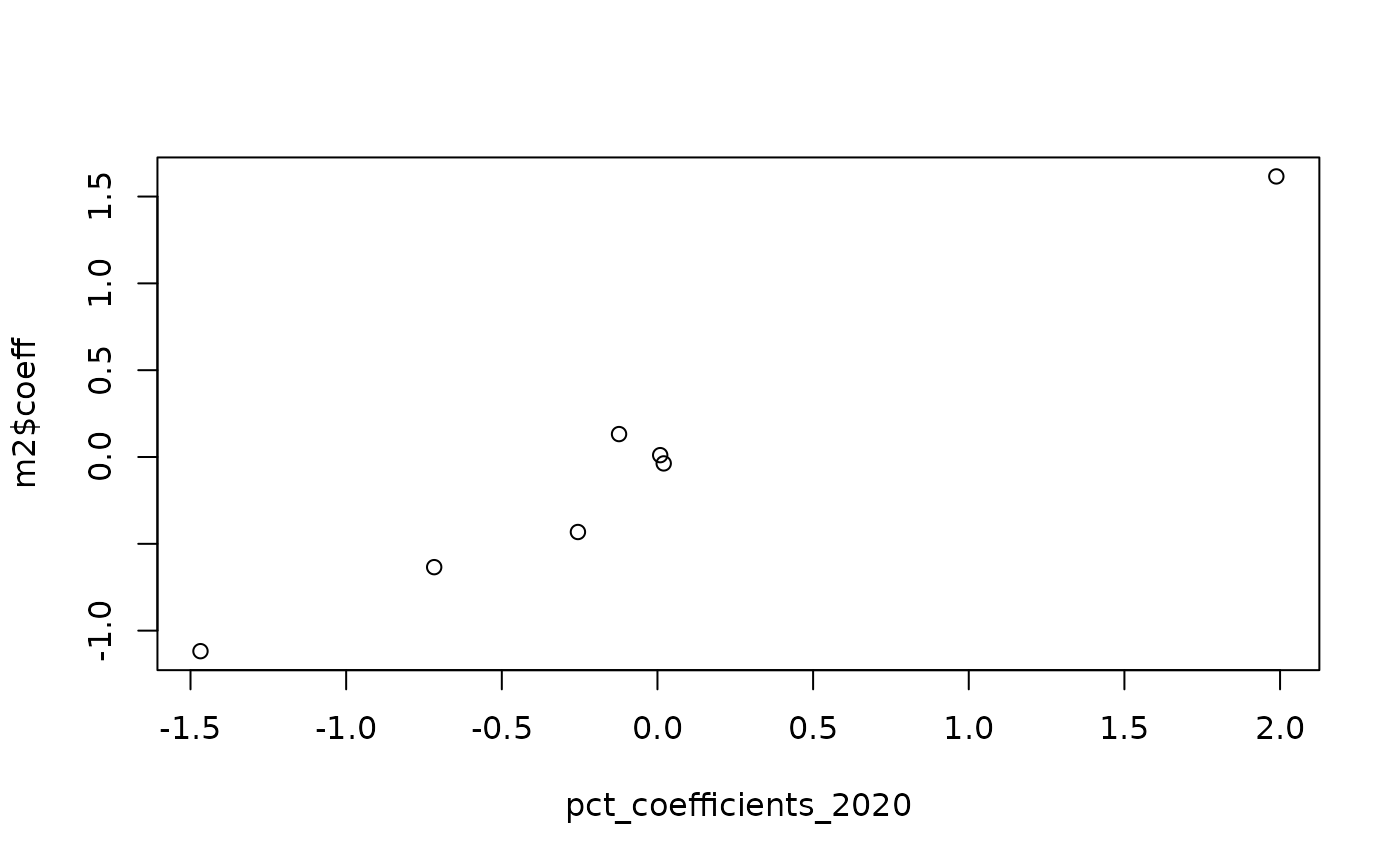 cor(pct_coefficients_2020, m2$coeff)^2
#> [1] 0.9740929
cor(pct_coefficients_2020, m3$coeff)^2 # explains 95%+ variability in params
#> [1] 0.9653234
cor(pct_coefficients_2020, m2$coeff)^2
#> [1] 0.9740929
cor(pct_coefficients_2020, m3$coeff)^2 # explains 95%+ variability in params
#> [1] 0.9653234